

Core logic of PyTOPKAPI.
The run function in this module contains the logic to run a TOPKAPI simulation based on the parameters specified in an INI file.
This module contains the routines used to solve the Ordinary Differential Equations in TOPKAPI.
Compute the final volume in a TOPKAPI water store for one time-step
This function calculates the volume in a generic store at the end
of the defined time-step, given the initial volume, inflow and ODE
parameters. This is done by solving the storage ODE 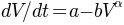 . Todini’s quasi-analytical solution can be
used as a speed up.
. Todini’s quasi-analytical solution can be
used as a speed up.
| Parameters: | I : scalar
b : scalar
alpha : scalar
Vt0 : scalar
Dt : scalar
solve_method : int
|
|---|---|
| Returns: | Vt1 : scalar
|
Quasi-analytical solution for volume in a TOPKAPI water store
This function calculates the volume in a generic store at the end
of the defined time-step, given the initial volume, inflow and ODE
parameters. This is done using Todini’s quasi-analytical solution
of the storage ODE 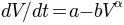 .
.
| Parameters: | a : scalar
b : scalar
alpha : scalar
V0 : scalar
Dt : scalar
|
|---|---|
| Returns: | V1 : scalar
|
Functions required by the TOPKAPI model for the management of input and output of cells.
Note
The subroutines solving the differential equation are not in this module (see ode for more information)
Compute the outflow Qout from a generic water store.
Calculate the mean outflow rate during the current time-step from a generic water store. The outflow is calculated as the difference between the inflow and rate of change in storage during the time-step.
| Parameters: | V_t0 : scalar
V_t1_prim : scalar
a : scalar
Dt : scalar
|
|---|---|
| Returns: | Qout : scalar
|
Compute the output flows Qout from the computed water volumes:
| Returns: | Qout : scalar
|
|---|
Partition the outflows from soil and overland stores.
Calculates the correct partitioning of outflows from the soil and overland stores into contributions going to the downstream cell and/or the channel store of the current cell (if this exists).
| Parameters: | Lambda : int
Qs_out : scalar
Qo_out : scalar
W : scalar
X : scalar
Xc : scalar
|
|---|---|
| Returns: | Q_to_next_cell : scalar
Q_to_channel : scalar
Q_to_channel_sub : scalar
|
Compute initial channel volume.
The initial volume ar_Vc_t0 is computed for all the channel cells, from a known initial flow.
Compute the total inflow to the channel of a channel cell.
Calculate the total inflow to the channel as the sum of inflows from upstream cells and inflows from the soil and overland stores in the current cell.
| Parameters: | ar_Qc_out : (N,) Numpy array
Q_to_channel : scalar
ar_cell_up : list of int
|
|---|---|
| Returns: | a_c : scalar
Qc_cell_up : scalar
|
Compute the total input to a soil store.
Calculate the total rate of input to a single soil store. This comprises the sum of rainfall input, subsurface contribution from upstream cells and the overland contribution from upstream cells.
| Parameters: | P : scalar
Dt : scalar
X : scalar
ar_Q_to_next_cell : (N,) Numpy array
ar_cell_up : list of int
|
|---|---|
| Returns: | a_s : scalar
|
Compute Manning depth for flow Q.
Compute with the manning equation (high width hypothesis) the water depth h corresponding to the flow Q for the channel characteristics: width W, manning n. The volume is then returned V=hWX
Compute the outflow from and volume in a soil store.
Calculate the outflow rate and final volume in a soil store after transferring volume greater than the saturated soil moisture content to the overland store of the same model cell.
| Parameters: | Vs_t0 : scalar
Vs_t1_prim : scalar
Vsm : scalar
a_s : scalar
b_s : scalar
alpha_s : scalar
Dt : scalar
|
|---|---|
| Returns: | Qs_out : scalar
Vs_out : scalar
|
Evaporation and evapotranspiration routines.
A selection of routines that can be used to compute the evapotranspiration losses in PyTOPKAPI.
Compute the evaporation from a channel store.
Calculate the evaporation loss from a channel store. The function attempts to remove the demand from the open water potential evaporation from the channel store, if there isn’t sufficient water in the store then the total volume in the channel is removed.
| Parameters: | Vc0 : scalar
ETo : scalar
W : scalar
X : scalar
|
|---|---|
| Returns: | ET_channel : scalar
Vc1 : scalar
|
Compute the evapotranspiration from a model cell.
The evapotranspiration loss is first taken from the overland store, if the storage in the overland store cannot satisfy the ET demand then the water is extracted from the soil store. In cases where the ET demand is greater than the available water, both the soil and overland store are totally drained.
| Parameters: | Vo0 : scalar
Vs0 : scalar
Vsm : scalar
kc : scalar
ETr : scalar
X : scalar
|
|---|---|
| Returns: | ETa : scalar
Vs1 : scalar
Vo1 : scalar
|
Functions required to compute the intrinsic TOPKAPI parameters from the physical parameters
Compute model parameters from physical parameters.
This function uses the physically based parameters and constraints for each model cell to compute, the saturated soil moisture volume, channel width and constant terms for the differential equations of the soil, overland and channel stores.
| Parameters: | X : scalar
ar_Xc : (N,) float array
Dt : scalar
alpha_s : scalar
alpha_o : scalar
alpha_c : scalar
nb_cell : scalar
A_thres : scalar
W_min : scalar
W_max : scalar
ar_lambda : (N,) int array
ar_tan_beta : (N,) float array
ar_tan_beta_channel : (N,) float array
ar_L : (N,) float array
ar_Ks : (N,) float array
ar_theta_r : (N,) float array
ar_theta_s : (N,) float array
ar_n_o : (N,) float array
ar_n_c : (N,) float array
ar_A_drained : (N,) float array
|
|---|---|
| Returns: | ar_Vsm : (N,) float array
ar_b_s : (N,) float array
ar_b_o : (N,) float array
ar_W : (N,) float array
ar_b_c : (N,) float array
|
Calculate the upstream cells for each cell.
This function calculates the immediate upstream cells contributing flow to each cell in the catchment.
| Parameters: | ar_cell_label : (N,) int array
ar_cell_down : (N,) int array
ar_label_sort : (N,) int array
|
|---|---|
| Returns: | li_cell_up : List of (M,) int arrays
|
Compute the drained area for each cell.
This function calculates the total area drained for each cell in the catchment, as the sum of it’s area and the upstream area.
| Parameters: | ar_label_sort : (N,) int array
li_cell_up : List of (M,) int arrays
X : scalar
|
|---|---|
| Returns: | ar_A_drained : (N,) float array
|
Read the spatially variable cell parameters from file.
Read the file containing the physical parameters of each cell. This information governs the distributed behaviour of the model.
| Parameters: | file_name : string
|
|---|---|
| Returns: | ar_cell_label : (N,) int array
ar_coorx : (N,) float array
ar_coory : (N,) float array
ar_lambda : (N,) int array
ar_Xc : (N,) float array
ar_dam : (N,) int array
ar_tan_beta : (N,) float array
ar_tan_beta_channel : (N,) float array
ar_L : (N,) float array
ar_Ks : (N,) float array
ar_theta_r : (N,) float array
ar_theta_s : (N,) float array
ar_n_o : (N,) float array
ar_n_c : (N,) float array
ar_cell_down : (N,) int array
ar_pVs_t0 : (N,) float array
ar_Vo_t0 : (N,) float array
ar_Qc_t0 : (N,) float array
ar_kc : (N,) float array
|
read_column_input
Read the file containing data in column format: Cell1 Cell2 Cell3 ... 1.3 4.3 5.2 ... 2.3 5.6 4.2 ...
Return a matrix mat_out(nrow, ncol)
Read global model parameters from file.
Read the file that specifies the parameters of the model, which are globally applied to all cells in the model.
| Parameters: | file_name : string
|
|---|---|
| Returns: | X : scalar
Dt : scalar
alpha_s : scalar
alpha_o : scalar
alpha_c : scalar
A_thres : scalar
W_min : scalar
W_max : scalar
|
Determine a suitable computation order for the cells.
Sort the cells into a valid computation order based on the distance to the outlet of the catchment. This is necessary as each cell depends on the contribution from upstream cells.
| Parameters: | ar_cell_label : (N,) int array
ar_cell_down : (N,) int array
|
|---|---|
| Returns: | ar_label_sort : (N,) int array
|
Find the label of the cell closest to (Xoutlet, Youtlet).
Find the label of the model cell containing the specified location. The co-ordinates of the location must be given in the same co-ordinate system as that specifying the model catchment.
| Parameters: | ar_cell_label : (N,) int array
Xoutlet : float
Youtlet : float
ar_coorx : (N,) float array
ar_coory : (N,) float array
ar_lambda : (N,) int array
channel : boolean (default=True)
|
|---|---|
| Returns: | cell_outlet : int
|
Utilities for reading and plotting ArcGIS binary files.
This module contains some simple functions to make it easier to read and plot the data contained in the binary grid files produced by ArcGIS.
Read the data field and headers from an ArcGIS binary grid
This function reads the header and data from the ArcGIS binary data files produced by the “Raster to Float” tool in ArcGIS 9.1
Read data from a ArcGIS binary file into an array.
Read the data from a binary file created by ArcGIS into a Numpy array. The file is expected to be in binary format with floating point precision. e.g. “.flt” extension.
Read the ascii headers of the ArcGIS binary grid file
The headers have the following format:
ncols 62 nrows 121 xllcorner -288595.47161281 yllcorner -3158065.5722693 cellsize 1000 NODATA_value -9999 byteorder LSBFIRST
 Copyright 2008-2010, Theo Vischel, Scott Sinclair & Geoff Pegram
Copyright 2008-2010, Theo Vischel, Scott Sinclair & Geoff Pegram
Last updated on Nov 01, 2010
| Created using Sphinx